OVERVIEW
OGM is a technology by which we can analyze structural and copy number variants at a high resolution through the evaluation of patterns generated by fluorophore tags labeled to specific sequence motifs within extremely long DNA molecules. It uses this ultra high molecular weight (HMW) DNA that are far longer than the DNA sequences analyzed in current second- and third-generation sequencing methods, to achieve average read lengths of over 200 kbp.
The optical genome mapping chip’s nanochannels allow only a single linearized DNA molecule to travel through while preventing the molecule from tangling or folding back on itself. Confined in this nanofluidic environment, DNA molecules linearize across hundreds of thousands of nanochannels where they can be imaged to reveal the underlying genomic structure and structural variation.
INDICATION
OGM is available for the following Indications:
TEST REQUIREMENTS
- Completed CGL Myeloid Testing requisition form
- Select Karyotpe/OGM adjacent to the appropriate hematological disorder
- Bone Marrow Aspirate (0.5mL fresh in EDTA tube) specimen (see Specimen Guidelines, DNA Molecular test type)
- Bone marrow report (reports can be faxed to CGL when finalized)
TURN-AROUND TIME
Approximately 14 days (AML) or 17-21 days (MDS, MPN) from receipt the final bone marrow report .
RESULTS REPORTING
- OGM reports include a list of variants classified into tiers of clinical significance
- TIER I – VARIANTS OF STRONG CLINICAL SIGNIFICANCE
- TIER II – VARIANTS OF POTENTIAL CLINICAL SIGNIFICANCE
- TIER IIIA –VARIANTS OF UNCERTAIN CLINICAL SIGNIFICANCE
- TIER IIIB – VARIANTS OF UNCERTAIN FUNCTION
- Please see our Variant Classification Guidelines for additional details.
Variants suspected to be of germline origin (constitutional) will be identified on the report as Potential Germline Findings, with an accompanying recommendation for referral for follow-up testing.
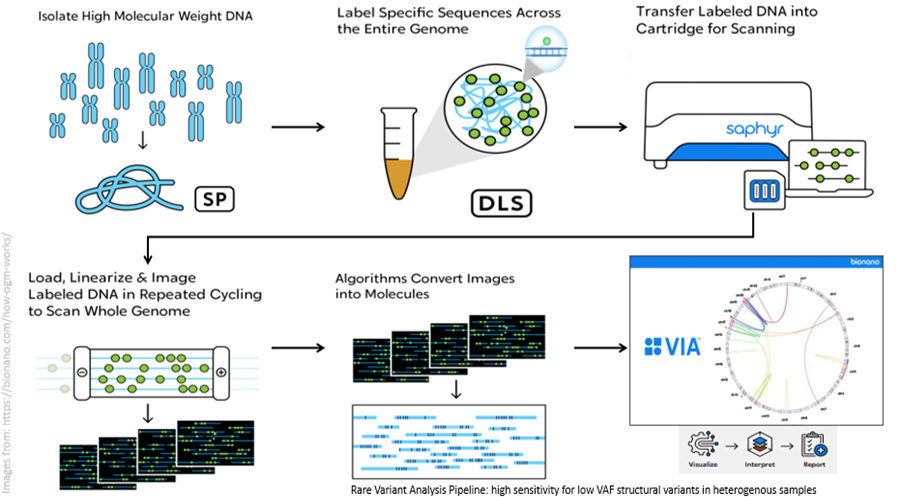
METHOD
Ultra-high molecular weight DNA is extracted from blood or bone marrow using the Bionano Prep SP-G2 Blood and Cell Culture DNA Isolation Kit. DNA is enzymatically labelled using the Bionano Prep DLS-G2 Labeling Kit according to the manufacturer’s protocol. Labeled DNA is loaded onto a nanochannel chip allowing the separation of single DNA molecules and imaged using the Bionano Saphyr Optical Genome Mapping instrument. Image files are converted into digital molecule maps and analysed using the Rare Variant Analysis (RVA) pipeline using GRCh38 human genome. Data is visualized using the Access Software Tool and variant annotation and filtering is performed with VIA for OGM data (Bionano Inc, San Diego, CA). Structural and numerical variants >5Mb are routinely reported. Structural and copy number variants <5Mb are filtered against a gene/region list that are known to have or with evidence supporting a diagnostic or prognostic significance in the malignancy being analysed (lists available upon request).
All filtered variants are tiered according to their known or predicted clinical significance (adapted from Levy et al PMID: 38164980 and Li et al. PMID:27993330). Tier I variants are those with a strong clinical significance and are predictive of response or resistance in a specific tumour type to therapies approved by Health Canada or US FDA. Alternatively, these variants are predictive, prognostic or diagnostic biomarkers in the specific tumour type, based on well-powered studies with consensus from experts in the field and/or inclusion in professional guidelines. Tier II variants are those with potential clinical significance are predictive, prognostic or diagnostic biomarkers, based on convincing published evidence from smaller studies or case reports but without expert consensus. Tier IIIA variants are those that have uncertain clinical significance and are known or presumed to alter normal gene function, however no convincing published evidence of a predictive, prognostic or diagnostic association was found or evidence is sufficiently conflicting that a conclusion cannot be reached. Tier IIIB variants are of uncertain function and are not observed at a significant allele frequency in population, pan-cancer or tumour-specific variant databases and their effect on normal gene function cannot be confidently predicted. No convincing published evidence of a predictive, prognostic or diagnostic association was found or evidence is sufficiently conflicting that a conclusion cannot be reached. Tier IV variants are known or presumed to not disrupt normal gene function, and/or are found at a significant allele frequency within the population and are not routinely reported.
OGM requires greater than 5-10% abnormal cells in a sample to reliably detect structural and copy number variants. This technique will not detect structural variation smaller than the lower limits of the analysis pipeline (including single nucleotide variants) – the minimum size for the rare variant assembly is ~5 Kb (5000 bases). OGM cannot detect any balanced whole-arm translocation that breaks in the repetitive DNA of the centromere, or higher states of ploidy (e.g. triploidy, tetraploidy) and has reduced sensitivity in repetitive regions such as the telomeres and centromeres. OGM detects the relative proportion of abnormalities in the sample but cannot separate independent clones (if present). This assay cannot reliably distinguish somatic (acquired) from constitutional (inherited) events.
Resources more information on this technology: